Chimera
Scripts
# open a protein and run chimera commands
from chimera import runCommand as rc # use 'rc' as shorthand for runCommand
from chimera import replyobj # for emitting status messages
replyobj.status('Reading 5nsr from pdb.org ...')
rc('open 5nsr')
rc('sel :.C :.D')
rc('del ~sel')
rc('sel :325.C :773.C :1161.C :361.D')
rc('color blue')
rc('color red sel')
IDLE
Tools -> General Controls -> IDLE
Protein
import chimera
# open a protein in Chimera
opened = chimera.openModels.open('3fx2', type="PDB") # a list of opened modules
mol = opened[0] # get the first molecule
# mol.name # pdf file name
# mol.atoms # atoms
# mol.bonds # bonds
# mol.residues # residues
# chimera.colorTable contains getColorByName and getTKColorByName
from chimera.colorTable import getColorByName
red = getColorByName('red') # _chimera.MaterialColor
blue = getColorByName('blue')
yellow = getColorByName('yellow')
mol.color = red # change model color to be red
atoms = mol.atoms
print(len(atoms))
for atom in atoms:
if atom.name == 'CA':
atom.color = yellow
else:
atom.color = blue
atom.drawMode = chimera.Atom.Ball
#coor = atom.xformCoord()
#print('%10s%10s\n' % (atom.name, atom.residue.id)) # disply stdout to IDLE
#print(coor.x, coor.y, coor.z)
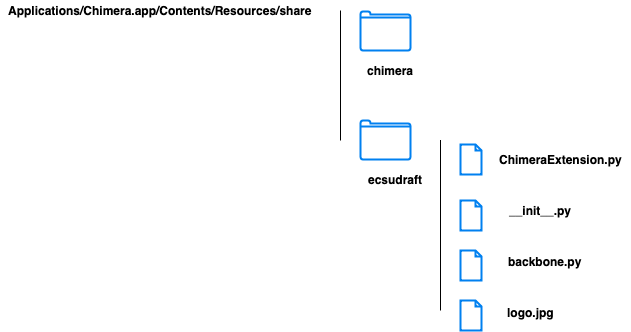
Extension
# ChimeraExtension.py
from chimera.extension import EMO, manager
# -----------------------------------------------------------------------------
#
class ModelZ_Dialog_EMO ( EMO ):
def name(self):
return 'ecsudraft' # extension name displayed in Chimera
def description(self):
return "a draft for ecsu validation tool" # description appeared as balloon help
def categories(self):
return ['Utilities'] # category in Chimera
def icon(self):
return self.path('logo.jpg') # logo
def activate(self):
# if no name is supplied, the "__init__.py" module is returned
self.module('backbone').colorBB() # locate module backbone and call its function colorBB()
# -----------------------------------------------------------------------------
# Register dialogs and menu entry.
#
manager.registerExtension ( ModelZ_Dialog_EMO ( __file__ ) )
# backbone.py
import chimera
from chimera import openModels, Molecule
from chimera.colorTable import getColorByName
def colorBB():
red = getColorByName('red')
yellow = getColorByName('yellow')
# return a list of opened molecules
for mol in openModels.list(modelTypes=[Molecule]):
mol.color = red # change model color to be red
atoms = mol.atoms
for atom in atoms:
if atom.name == 'CA':
atom.color = yellow # change CA atoms to be yellow
Install Biopython
Download biopython
python setup.py install --home=/Applications/Chimera.app/Contents/Frameworks/Python.framework/Versions/2.7/
Enter /Applications/Chimera.app/Contents/Frameworks/Python.framework/Versions/2.7/lib, "mv ./python/* ./python2.7", "rm -r ./python", # the default install directory is /python instead of /python2.7
Restart Chimera, open IDLE, type "import Bio"
Reference