Biopython
PDB IO
# Read proteins from PDB files
#!/usr/bin/python
from Bio.PDB import *
import numpy as np
def getStructure(fileName):
# create a protein structure parser
parser = PDBParser();
# read the protein structure with the parser
try:
structure = parser.get_structure('5szs', fileName);
return structure;
except Exception, err:
print err
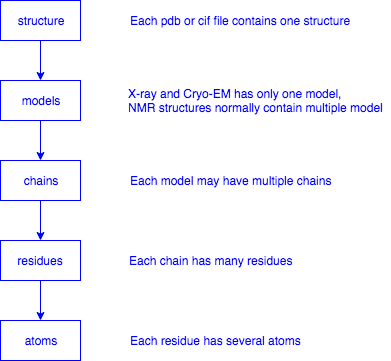
def getModels(structure):
for model in structure.get_models():
yield model;
def getChains(model):
for chain in model.get_chains():
yield chain;
def getResidues(chain):
for residue in chain.get_residues():
yield residue;
def getAtoms(residue):
for atom in residue.get_atoms():
yield atom;
def getAtomInfo(atom):
print atom.get_name(), atom.get_id(), atom.get_full_id(), atom.get_coord(), atom.get_bfactor(), atom.get_occupancy(), atom.get_fullname(), atom.get_altloc(), atom.get_vector(), atom.get_vector().get_array(), atom.get_parent().get_id()
# Walk through protein with iteration
def getStructure(fileName):
# create a protein structure parser
parser = PDBParser();
# read the protein structure with the parser
try:
structure = parser.get_structure('5szs', fileName);
return structure;
except Exception, err:
print err
def main():
structure = getStructure('4mv2.pdb');
for model in structure:
for chain in model:
for residue in chain:
print residue.get_resname()
# Read proteins from cif files
def getStructure(fileName):
# create a protein structure parser
parser = MMCIFParser();
# read the protein structure with the parser
try:
structure = parser.get_structure('5szs', fileName);
return structure;
except Exception, err:
print err
# Output a structure to a pdb file
def getStructure(fileName):
# create a protein structure parser
parser = PDBParser();
# read the protein structure with the parser
try:
structure = parser.get_structure('5szs', fileName);
return structure;
except Exception, err:
print err
def main():
''' Generate dihedral angles for a specific residue
Args:
residueName (string), target residue
'''
structure = getStructure('5szs.pdb');
# Dice
Dice.extract(structure, 'A', 1, 100, 'temp2.pdb');
# PDBIO
io = PDBIO();
sel = Dice.ChainSelector('A', 1, 100);
io.set_structure(structure);
io.save('temp.pdb', sel);
Download PDB
def getStructure(fileName):
pdbl = PDBList();
pdbl.retrieve_pdb_file(fileName, file_format = 'mmCif');
def main():
''' Generate dihedral angles for a specific residue
Args:
residueName (string), target residue
'''
structure = getStructure('1FAT');
PDB Header
# Read header information from a pdb or cif file
def getStructure(fileName):
# create a protein structure parser
parser = PDBParser();
# read the protein structure with the parser
try:
structure = parser.get_structure('5szs', fileName);
return structure;
except Exception, err:
print err
def main():
''' Generate dihedral angles for a specific residue
Args:
residueName (string), target residue
'''
structure = getStructure('5szs.pdb');
print structure.header;
Reference